TUTORIAL
Long non-coding RNA network
The Me-lncRNAs was constructed by using cassava genome version 6.1 (Phytozome version 12.0) based on integrating of comparative- and transcriptome- based approach. Six plant genomes; Arabidopsis (Arabidopsis thaliana), Poplar (Populus trichocarpa), Castor bean (Ricinus communis), Jatropha (Jatropha curcas) and 2 cassava cultivars (W14 and KU50) were retrieved from plant genome databases for ncRNAs prediction by using comparative-based algorithm, RNAz. Seventy-four cassava RNA-seq datasets generated from 5 publications and previously published ncRNAs in RNAcentral, NCBI, GreeNC, CANTATAdb, miRbase and Rfam were used for verification. Finally, novel ncRNAs with expression supporting and passed through the filtering criteria were designated as Me-lncRNAs. Expression evidence of Me-lncRNAs indicates by number of publications supporting the expression. Of these with differential expression under cold and/or drought stress were predicted their targets based on cis- or trans- regulation.
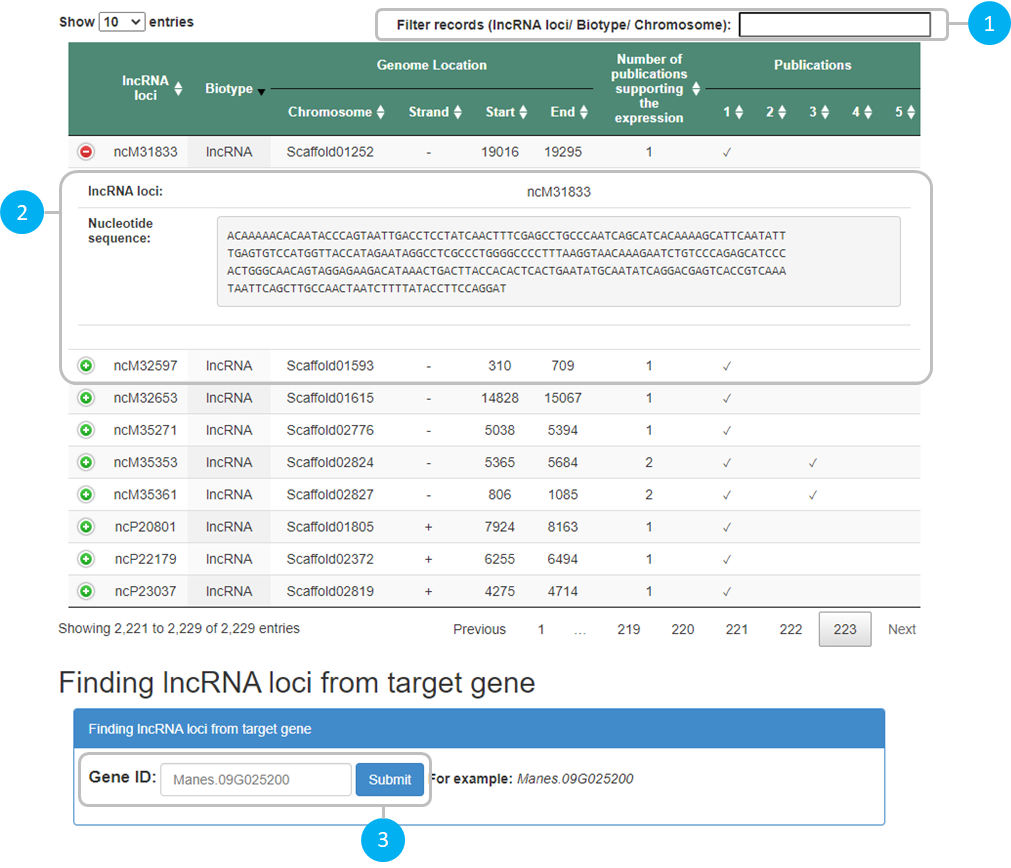
1 |
Searching lncRNA loci and location |
|---|---|
2 |
Displaying information of particular lncRNA loci |
3 |
Finding lncRNA loci from protein-coding gene target |
Description of terminology
| lncRNA loci | : ID of putative lncRNAs | |
|---|---|---|
| Biotype | : Types of putative lncRNAs based on genomic location including (a) lincRNA, (b) lncNAT and (c) sense/intronic lncRNA | |
| - lincRNA | : LncRNA locating at intergenic region without protein coding genes overlapping | |
| - lncNAT | : LncRNA locating at intergenic region with protein coding genes overlapping in the opposite strand | |
| - Sense/intronic lncRNA | : LncRNA located at untranslated region or intron of protein coding gene | |
| Genome Location | : Location of putative lncRNAs in cassava genome version 6.1 (i.e., chromosome, scaffold) based on Phytozome version 12 | |
| - Chromosome | : Chromosome in cassava genome version 6.1 based on Phytozome version 12 | |
| - Strand | : DNA strand (sense (+) or antisense (-)) in cassava genome version 6.1 based on Phytozome version 12 | |
| - Start | : Started position of putative lncRNA in cassava genome version 6.1 based on Phytozome version 12 | |
| - End | : End position of putative lncRNA in cassava genome version 6.1 based on Phytozome version 12 | |
| Nucleotide sequence | DNA sequences of putative lncRNAs | |
| Number of publications supporting the expression | : Number of supporting putative lncRNA expression evidence (maximum is 5) | |
| Publications | : Found cassava RNA-seq datasets supporting the expression of cassava lncRNAs | |
| Relationship | : Relationship of putative lncRNA and its target genes based on cis- or trans- regulation | |
| - Cis-regulation | : Target prediction based on genomic location of protein coding gene located within 10 Kb. in up or downstream of putative lncRNA | |
| - Trans-regulation 1 | : Target prediction based on sequence complementary and normalized binding free energy (ndG) < -0.1 of putative lncRNAs and mRNA of protein coding gene target using LncTar | |
| - Trans-regulation 2 | : Target prediction based on the same mature miRNA binding with mismatch ≤ 2 of putative lncRNA and mRNA of protein coding gene target using psRNATarget | |
| - Target gene ID | : Target gene ID in cassava genome version 6.1 based on Phytozome version 12 | |
| - Condition | : Condition which was found relationship between putative lncRNA and target gene including cold and/or drought stress | |
| - miRNA | : miRNA which was found to share binding region on putative lncRNA sequence and mRNA sequence of target gene |